The latest Drosophila melanogaster 5 genome has a complete copy of the mitochondrial chromosome embedded in chrU, which makes it a trap for mitochondrial RNAseq reads. If the aligner randomly distributes reads between all matching sites, half of the mitochondrial reads will go to chrU. All reads will be lost if ambiguous alignments are discarded.

Peak: mitochondrial reads aligned to chrU. (UCSC genome browser)
ChrU is not a real chromosome, but a 10 Mb mixed bag of 34,630 small scaffolds, that didn’t seem to fit anywhere during shotgun genome assembly. My best guess is, that the 19.5 kb mitochondrial scaffold seemed far too small to be anything real during genome assembly, so the Celera shotgun assembler just lumped it into chrU with all the other loose fragments. According to this paper, the fragment is the true y1, cn1, bw1, sp1 strain mitochondrial genome, in contrast to the reference chrM, which is a composite of several genomes.
Leaving out all, or parts of chrU during alignment is an obvious solution. The coordinates of the mitochondrial bit are (roughly) chrU:5288508-5303826, and turning this stretch into N’s preserves the remainder of chrU, without trapping mitochondrial reads. Another option is to leave out chrU altogether. This is probably justifiable given that most of chrU are just duplicated fragments from other parts of the genome, only sequenced in much worse quality and thus not fitting into their original place.
[Update] I heard BDGP will remove this snag in the release 6 genome.
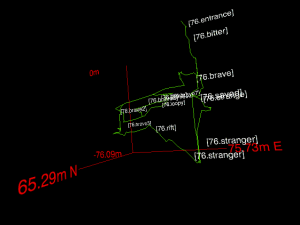 Check out this small survey or this cave system (this one might crash browsers on older machines). The controls are as usual: drag for rotating, scroll for zoom. The surveys were made by the Cambridge University Caving Club (CUCC)‘s Austria expeditions, and over time, Squiggle will be used to preview them. Thanks to Wookey for his help in making and testing Squiggle.
Check out this small survey or this cave system (this one might crash browsers on older machines). The controls are as usual: drag for rotating, scroll for zoom. The surveys were made by the Cambridge University Caving Club (CUCC)‘s Austria expeditions, and over time, Squiggle will be used to preview them. Thanks to Wookey for his help in making and testing Squiggle.![David Molnar [Update:, PhD]](https://www.srcf.ucam.org/~dm516/wp-content/themes/twentyeleven/images/headers/chessboard.jpg)
